How Small Protein Fragments Can Unlock the Secrets of Protein Interactions
All biological function is dependent on how different proteins interact with each other. Protein-protein interactions facilitate everything from transcribing DNA and controlling cell division to higher-level functions in complex organisms. However, much remains unclear about how these functions are orchestrated on the molecular level, and how proteins interact with each other – either with other proteins or with copies of themselves.
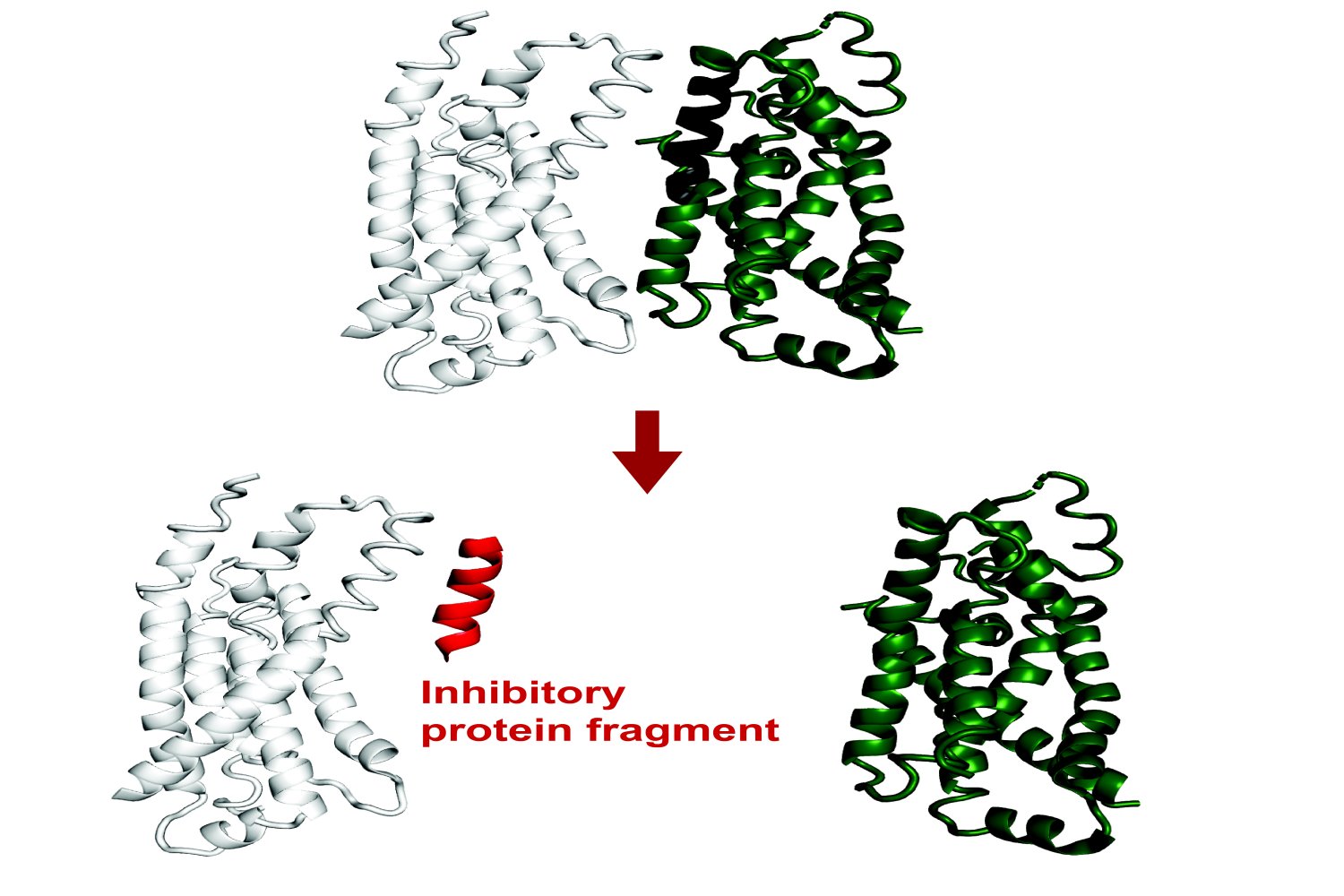
Recent findings have revealed that small protein fragments have a lot of functional potential. Even though they are incomplete pieces, short stretches of amino acids can still bind to interfaces of a target protein, recapitulating native interactions. Through this process, they can alter that protein’s function or disrupt its interactions with other proteins. Protein fragments could therefore empower both basic research on protein interactions and cellular processes, and could potentially have therapeutic applications.
A New Method for Predicting Protein Fragments
Recently, a new method was developed in the Department of Biology to computationally predict protein fragments that can bind to and inhibit full-length proteins in E. coli. Theoretically, this tool could lead to genetically encodable inhibitors against any protein. The work was done in the lab of associate professor of biology and Howard Hughes Medical Institute investigator Gene-Wei Li in collaboration with the lab of Jay A. Stein (1968) Professor of Biology, professor of biological engineering, and department head Amy Keating.
Leveraging Machine Learning
The program, called FragFold, leverages AlphaFold, an AI model that has led to phenomenal advancements in biology in recent years due to its ability to predict protein folding and protein interactions. The goal of the project was to predict fragment inhibitors, which is a novel application of AlphaFold. The researchers on this project confirmed experimentally that more than half of FragFold’s predictions for binding or inhibition were accurate, even when researchers had no previous structural data on the mechanisms of those interactions.
Inhibition, and Beyond
The researchers accomplished these predictions by computationally fragmenting each protein and then modeling how those fragments would bind to interaction partners they thought were relevant. They compared the maps of predicted binding across the entire sequence to the effects of those same fragments in living cells, determined using high-throughput experimental measurements in which millions of cells each produce one type of protein fragment.
Design, in Principle
This research is a starting point for developing a systemic understanding of cellular design principles, and what elements deep-learning models may be drawing on to make accurate predictions. "There’s a broader, further-reaching goal that we’re building towards," Savinov says. "Now that we can predict them, can we use the data we have from predictions and experiments to pull out the salient features to figure out what AlphaFold has actually learned about what makes a good inhibitor?"
Conclusion
This research has the potential to revolutionize our understanding of protein interactions and their role in biological processes. The development of FragFold, a new method for predicting protein fragments that can bind to and inhibit full-length proteins, is a significant step forward in this area. The ability to predict fragment inhibitors with high accuracy and precision opens up new possibilities for manipulating protein function and could have important implications for a range of fields, from basic research to medicine.
Frequently Asked Questions
Q: What is FragFold?
A: FragFold is a new method for predicting protein fragments that can bind to and inhibit full-length proteins.
Q: How does FragFold work?
A: FragFold leverages AlphaFold, an AI model that predicts protein folding and protein interactions, to predict fragment inhibitors.
Q: What are the potential applications of FragFold?
A: The potential applications of FragFold are vast, from basic research to medicine. It could be used to develop new treatments for diseases, to study protein interactions, and to manipulate protein function.
Q: Is FragFold limited to predicting inhibitor fragments?
A: No, FragFold is not limited to predicting inhibitor fragments. It can predict any type of fragment that binds to a target protein, including fragments that can stabilize the protein, enhance or alter its function, or trigger protein degradation.